DNA sequencing can be carried out in a number of different ways depending on the needs of an experiment. The ONT MinION sequencing device offers a portable, versatile, efficient, and relatively inexpensive way to sequence DNA in a classroom setting. In order to sequence DNA amplicons on this platform, they must first be prepared using an ONT kit. One efficient option for amplicons is the Rapid Barcoding Kit.
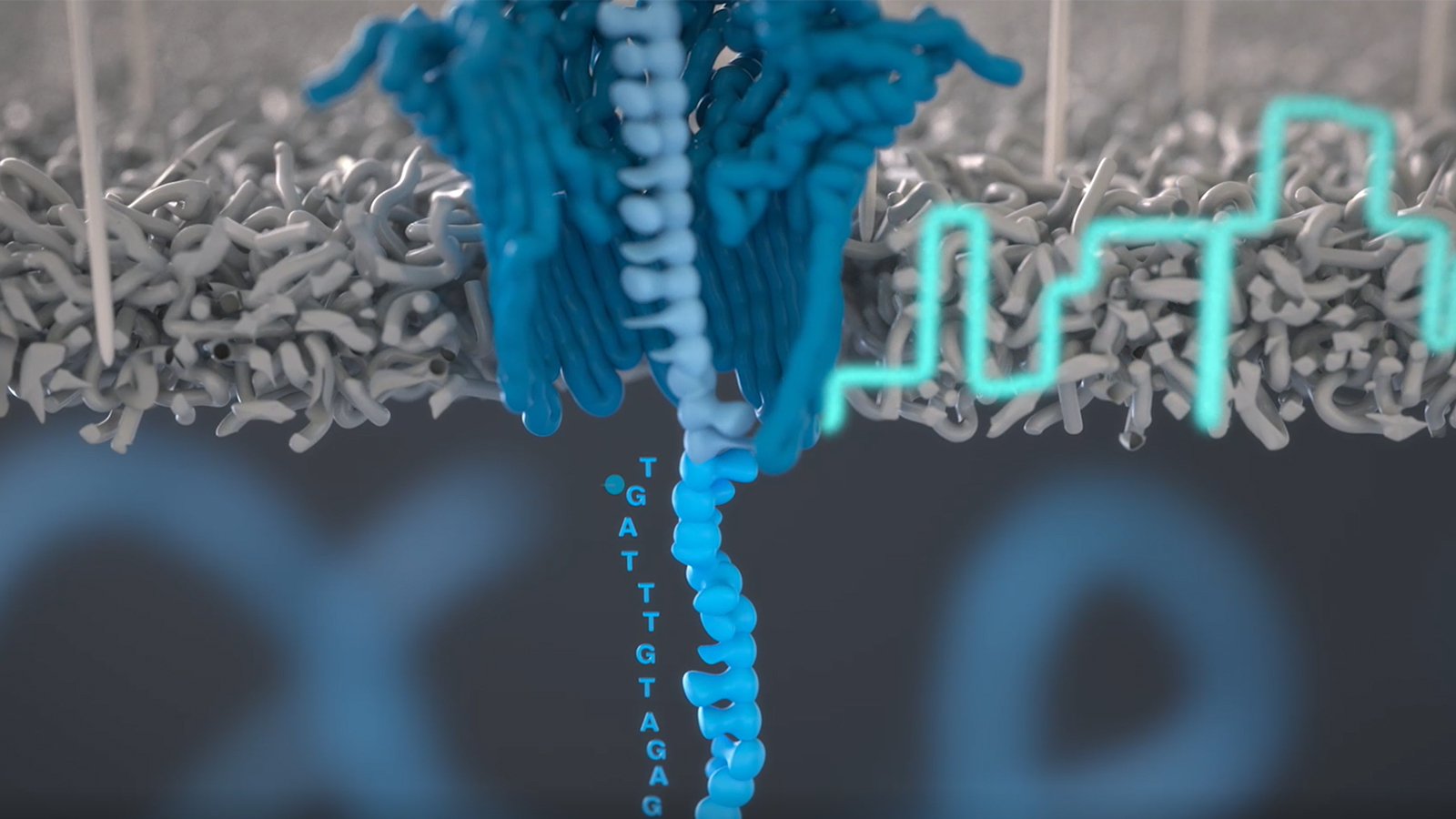
The ONT Rapid Barcoding Kit utilizes a transposome complex to cut and modify DNA. The transposome consists of a transposase enzyme and a DNA molecule. In nature, transposases can cut out DNA sequences called transposons and insert them into a new location within a genome. In the ONT kit, the transposase cuts double-stranded DNA that is to be sequenced, and then splices a small piece of DNA onto the ends of the cleaved DNA fragments. The DNA molecule that is attached to DNA fragments contains ONT “barcodes.” These barcodes are like “nucleotide name tags” that identify all DNA fragments associated with a sample. In other words, they are oligonucleotide indexes, which tag all the DNA from a sample so that multiple samples can be sequenced at once. In addition, the barcode indexes also have chemical groups on the ends of each sequence that will allow for the addition of sequencing adapters in a subsequent step. The sequencing adapters contain an additional DNA sequence and a motor protein with helicase activity. Together, they facilitate the movement of DNA sequences to a nanopore protein and their subsequent sequencing. Nanopore and motor proteins are at the heart of ONT technology, as they will actually perform the work of DNA sequencing.
ONT DNA sequencing is carried out by feeding single-stranded DNA (ssDNA) molecules through a series of nanopores embedded in an artificial membrane. This membrane exists within the environment of a flow cell and is bathed in an ionic solution, such that ions flow through the nanopores and elicit an electrochemical potential difference across the membrane. Once DNA is brought near a nanopore on one side of the membrane, it is coordinated to the mouth of the pore via a tether molecule. The sequencing adaptor DNA sequence is recognized by the mouth of the nanopore and the motor protein will unwind and feed ssDNA molecules through the pore. As ssDNA moves through the pore, the physical presence of nucleotides within the channel of the nanopore will impede the flow of ionic current through the pore, perturbing the electrochemical potential across the pore. This occurs in a characteristic way depending on the combination of nucleotides in the sequence. ONT’s sequencing devices, the MinION, capture this electrical data and write it to a file format called POD5. In a process called “basecalling”, the electrical signals are assigned a series of nucleotides and returned to the user as a series of FASTQ files for bioinformatics analyses.
This protocol is designed for educators to guide students through an optional PCR clean-up of up to 24 DNA amplicons and attachment of ONT Rapid Barcodes from the Rapid Barcoding Kit (product number: SQK-RBK114.24), the initial steps of DNA amplicon sequencing. It also walks educators through pooling of DNA amplicons, clean-up of the amplicon library, library preparation, and priming and loading of a flongle flow cell (FLG-FLO114). Note that a flongle flow cell is our recommended platform for amplicon sequencing for educators. This protocol assumes that the user has already conducted DNA extraction, PCR amplification, and gel electrophoresis confirmation of PCR success.
