Symposium: 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012
2024-25 Symposium
Thank you to all of the teams who submitted proposals, performed experiments, and presented project results at the poster sessions held at The City Tech Theater on Tuesday, May 20, 2025
At a glance:
- 157 students
- 17 mentors
- 15 schools and institutions
51 DNA barcoding projects:
- 37% plant
- 37% invertebrate
- 14% fungi
- 9% bacteria
- 4% lichen
- 4% vertebrate
Keynote Address
From Microbes to Megafauna: The Role of Metabarcoding in Shaping Oceanography
Elizabeth Suter, Ph.D.
Associate Professor of Environmental Science
Molloy University
Outstanding Poster
Mimicry Complexes and Host Plant Species of Seed Bugs on Randall’s Island
Meredith Metz, Mia Seshadri, and Rose Pasternak
Mentored by Abigale Kappa
Ethical Culture Fieldston School, The Bronx
In this experiment, we plan to collect three species from the family Lygaeidae: Lygaeus turcicus, the false milkweed bug (FMB), Lygaeus kalmii, the small milkweed bug (SMB), and Oncopeltus fasciatus, the large milkweed bug (LMB). Our purpose is to determine the impact of the habitat type and host plant species on the number of each milkweed bug species found on Randall’s Island. Additionally, we aim to study Batesian mimicry (the FMB is the mimic and the SMB is the model). Our research question is: How do the habitat type and host plant species affect the number of small, large, and false milkweed bugs we will find on Randall’s Island? To do this, we will collect these species from three habitats (Freshwater Meadow, Freshwater Wetland, and Little Hell Gate Salt Marsh) on Randall’s Island, isolate the DNA, amplify it using PCR, and analyze it through gel electrophoresis. After the DNA is extracted, amplified, and analyzed, we will create a graph depicting the number of each milkwe
2023-24 Symposium
Thank you to all of the teams who submitted proposals, performed experiments, and presented project results at the poster sessions held at The City Tech Theater on Wednesday, May 29, 2024 from 4:00-7:00 PM.
At a glance:
- 180 students
- 19 mentors
- 15 schools
55 DNA barcoding projects:
- 44% plants
- 27% invertebrates
- 16% fungi
- 9% bacteria
- 2% lichen
Keynote Address
DNA Barcoding, oh the places you'll go!
Stephen E. Harris, Ph.D.
Assistant Professor of Biology
Purchase College, SUNY
Outstanding Poster
The Effect of Invasive Stiltgrass on Biodiversity of Ground-Level Plant Species in Northern NJ Forests
Wendy Guo and Hillary Xie, Tenafly High School
Mentored by Anat Firnberg, Tenafly High School
Japanese Stiltgrass reduces biodiversity of ground-level plant species due to its smothering growth and potentially negative effects on soil composition. We studied its effects on local ecosystems by examining plant-life and soil composition in both stiltgrass-infested and control, or stiltgrass-free areas. We collected soil and plant tissue samples from various plots in forested areas. Infested plots hosted lower biodiversity in terms of species richness and relative abundance. We conclude that Japanese Stiltgrass reduced biodiversity drastically and dominated the forest floor by amassing in dense patches, leaving little room for competitor species. Stiltgrass outcompetes less resilient native species and destabilizes the ecosystem, disrupting normal ecological function. Our soil tests have indicated that Stiltgrass growth raises phosphorus levels but does not affect soil composition significantly otherwise.
2022-23 Symposium
Thank you to all of the teams who submitted proposals, performed experiments, and presented project results at the poster sessions held at The City Tech Theater on Wednesday, May 31, 2023 from 4:00-7:00 PM.
At a glance:
- 124 students
- 20 mentors
- 18 schools
45 DNA barcoding projects:
- 42% plants
- 33% invertebrates
- 18% lichen
- 7% fungi
Keynote Address
Autocatalytic Mutualisms and the Evolution of Biological Systems
David Kizirian, Ph.D.
Curatorial Associate, Department of Herpetology, Division of Vertebrate Zoology
American Museum of Natural History
Outstanding Poster
Biodiversity of Fungi in Northern New Jersey
Justin Byun and Nikhil Jathavedam, Tenafly High School
Mentored by Anat Firnberg, Tenafly High School
The fungi kingdom acts as decomposers, using enzymes to break down organic materials, and as networkers of nutrients, spreading wide networks called mycelium that interacts with plant roots forming symbiotic relationships. The study identified the species of fungi and estimate fungi density in Northern New Jersey that have implications for preserving forest habitats in rapidly changing environments due to pollutants. Results from soil testing revealed that pH soil levels were predominantly acidic to neutral, and the nitrogen and phosphorus levels were adequate to surplus. DNA analysis and density calculations revealed high fungal biodiversity in forests and low amounts of fruiting bodies. The numerous species of fungi in Northern New Jersey harbor an advantageous environment for the growth of plants that are essential for the health of forest habitats.
2021-22 Symposium
Thank you to all of the teams who submitted proposals, performed experiments, and presented project results at the poster sessions held at The City Tech Theater on Friday June 3, 2022 from 4:00-7:00 PM.
At a glance:
- 104 students
- 18 mentors
- 15 high schools
37 DNA barcoding projects:
- 59% animals
- 38% plants
- 3% lichen
Keynote Address
Disturbed and Diverse: NYC microbial communities have elevated levels of biodiversity
Theodore Muth, Ph.D.
Professor, CUNY Brooklyn College
Outstanding Poster
Performing Barcoding and Soil Analysis to Investigate Ecosystem Resilience in Different Flood Zones of Brooklyn, NY
Veronica Ng and Eileen Zheng, Brooklyn Technical High School
Mentored by Michael Estrella and Risa Parlo, Brooklyn Technical High School
With climate change and rising sea levels, regions near the coast, including south Brooklyn, are exposed to massive flooding and intense precipitation. Previous research discovered flooding shifts species distribution and soil acidity and salt concentrations increase while potassium (K), nitrogen (N) and phosphorus (P) levels decrease. We therefore predicted a trend in the evacuation zones that would go from low pH-high salt-low nutrient in more flood prone areas to high pH-low salt-high nutrient in less flood prone areas. Thus, areas more susceptible to flooding would have greater salt-tolerant species. Surprisingly, after DNA barcoding and performing soil analyses on samples collected from every zone, we found no such correlation between the evacuation zone and pH, salt concentration, and nutrient levels. Ultimately, investigating if there is a correlation between evacuation zones and the prevalence of different species will hopefully assist in guiding solutions to weathering climate change and protecting biodiversity.
2020-21 Symposium
Thanks to all of the teams who submitted proposals, performed experiments, and presented project results at the Virtual Symposium on Wednesday, June 1, 2021.
At a glance:
- 33 participating teams
- 96 students
- 16 mentors
Keynote Address
Understanding, deconstructing and rebuilding microbiomes to make a better world
Javier A. Izquierdo, Ph.D.
Associate Professor of Biology, Hofstra University
2019-20 Symposium
The ninth year of the Urban Barcode Project concluded on Thursday, June 4, 2020 during a Virtual Symposium.
At a glance:
- 27 participating teams
- 116 students
- 12 mentors
Keynote Address
How I became a rainforest explorer: Ant genomes to microbiomes
Corrie S. Moreau, Ph.D.
Moser Professor of Arthropod Biosystematics and Biodiversity and curator of the Cornell University Insect Collection (CUIC)
2018-19 Symposium
The eighth year of the Urban Barcode Project concluded on May 30, 2019 with poster presentations at the New York Academy of Medicine, New York.
At a glance:
- 41 participating teams
- 119 students
- 17 mentors
- 12 high schools and 1 academic institution
Taxonomic group studied:
- 59% animals
- 29% plants
- 7% eDNA
- 2% lichen
- 2% fungi
Students collected 600+ samples—an average of 15 samples per team—that generated over 630 sequences.
Keynote Address

Wild Microbes
Claudia Wultsch, Ph.D.
Hunter College, City University of New York
Sackler Lab of Comparative Genomics, American Museum of Natural History
Jaguar Program, Panthera
Outstanding Poster
Investigating the Origins of Invasive Earthworms in the Soil of Public, Private, Residential, and Commercial Areas of Greenpoint
Katya Naphtali, Institute for Collaborative Education, Manhattan
Mentored by TR Muth, Brooklyn College
Earthworms species diversity indicates pollution levels and abundance indicates disruption of soil structure. The student utilized earthworms as bioindicators in public and private locations in Greenpoint, Brooklyn to observe the functionality of the soil and need for bioremediation. More than 30 earthworms from 12 sites were collected, identified taxonomically, and identified genetically through DNA Barcoding. The abundance data demonstrates no significant difference between public and private locations. Based on DNA sequencing, the diversity is higher in private locations than public locations, indicating that they are less disturbed.
2017-18 Symposium
The seventh year of the Urban Barcode Project concluded on May 24, 2018 with poster presentations at the New York Academy of Medicine, New York. Supported by The Thompson Family Foundation.
At a glance:
- 54 participating teams
- 160 students
- 22 mentors
- 19 high schools
Taxonomic group studied:
- 54% animals
- 38% plants
- 4% fungi
- 4% other
Students collected 820+ samples—an average of 15 samples per team—that generated over 700 sequences.
Keynote Address
An Ecosystem in a Drop of Water: Molecular Approaches to Surveying Aquatic Biodiversity in Urban Habitats.
Elizabeth Alter, Ph.D.
Assistant Professor
Marine Evolutionary & Ecological Genetics
York College and The Graduate Center, CUNY
Outstanding Poster
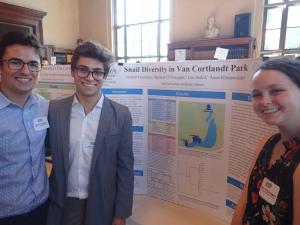
Snail Diversity in Van Cortlandt Park
Garrett Friedman, Leo Satlof, and Spencer O'Flanagan.
Mentored by Anne Kloimwieder, Ethical Culture Fieldston School
The winning team examined the relationship between water quality at various locations of Van Cortlandt Park and the species of snail present. Students collected more than 30 snails from three separate locations within the park and identified 6 different species through DNA barcoding. At each collection, site water quality measurements were recorded, as high fertilizer runoff and pollution affect the species that reside in the water. Results indicate that all locations investigated contained many more pollution-resistant lunged snails than pollution-sensitive gilled snails.
2016-17 Symposium
The sixth year of the Urban Barcode Project concluded on May 25, 2017 with poster sessions at the New York Academy of Medicine, New York. Funded in part by The Thompson Family Foundation
At a glance:
- 43 participating teams
- 139 students
- 21 mentors
- 18 high schools
Taxonomic group studied:
- 56% plants
- 21% animals
- 14% other
- 9% microbes
Students collected 570+ samples—an average of 13 samples per team—that generated over 900 sequences.
Keynote Address
Some Adventures in Urban Barcoding
Jesse H. Ausubel
Director, Program for the Human Environment
The Rockefeller University
Outstanding Posters
Barcoding Roots to Identify Biodiversity at the Margin of a Small Urban Forest
Nathalee Almonte, Julia Martino, and Jesus Bailon, Hostos-Lincoln Academy.
Mentored by Allison Granberry, Hostos-Lincoln Academy, and Damon P. Little, New York Botanical Garden.
The winning team studied the biodiversity of a small urban forest, the Thain Family Forest in the Bronx, New York, by DNA barcoding individual roots found in soil samples. The results were compared to an “above ground” morphological data bank in order to determine if below ground plant material could characterize the above ground ecosystem. Results show that while five plant species were reliably identified, DNA barcoding roots did not resolve several other samples to the species level. More than one fourth of the species collected were an invasive species, the Eurasisan native Poa annua.
2015-16 Symposium
The fifth year of the Urban Barcode Project concluded on June 3, 2016 with poster sessions at the Borough of Manhattan Community College, New York.
At a glance:
- 57 participating teams
- 172 students
- 27 mentors
- 22 high schools
Taxonomic group studied:
- 44% plants
- 37% animals
- 10.5% algae
- 7% fungi
- 1.5% other
Students submitted 850+ samples for sequencing—an average of 15 samples/team —that generated over 1200+ sequences.
Outstanding Posters
Management of an Invasive Species, Corydalis incisa, Along the Bronx River
Sammy Baez, Rosa Bermejo, Josiah Estacio and Sydia Fraguada, mentored by Allison Granberry, Hostos Lincoln Academy, and Damon Little, New York Botanical Garden
Assessing the Accuracy of the Usage of Lichens as Index Organisms Through the Assessment of Lichen Species with DNA Barcoding
Bryant Lee, Jai Yoon Chung, and Kengo Kuwama, mentored by Helen Coyle, Tenafly High School
The Effect of Salt Marsh Restoration on Ant Biodiversity
Akansha Thakur, Ilona Petrychyn, and Indu Puthenkalam, mentored by Camila Lock, Forest Hills High School
2014-15 Symposium
Thanks to all of the teams who submitted proposals, performed experiments, and presented project results at the poster sessions held at the American Museum of Natural History on May 27, 2015. Seven teams were selected to give oral presentations and compete for the grand prize at the UBP Symposium Award Ceremony on June 8, 2015 at AMNH.
At a glance:
- 50 participating teams
- 170 students
- 29 mentors
- 22 high schools, 14 public and 8 private
Taxonomic group studied:
- 62% plants
- 29% animals
- 7% microbes
- 2% fungi
Students submitted 700+ samples for sequencing—an average of 14 samples/team —that generated over 900+ sequences.
Keynote & Finalist Presentations
- Welcome:
[00:00:00] - Award Ceremony Remarks:
[00:03:30] Keynote – Susan Perkins, Ph.D. - Finalist Presentations:
[00:45:00] Proof of Panax
[00:55:40] Fish Fraud in NYC
[01:07:00] In Search of Invasive Plants
[01:18:45] Bacteria in NY Water Sources
[01:31:00] Smartphone Bacterial Microbiomes
[01:41:20] The Impacts on a Marine Ecosystem
[01:53:30] Macroinvertebrate Bioindicators
Awards Ceremony
Judges:
- Susan Perkins, Ph.D.
Curator and Professor, Sackler Institute for Comparative Genomics and Division of Invertebrate Zoology, American Museum of Natural History - Ana Fernandez-Sesma, Ph.D.
Associate Professor of Microbiology, Icahn School of Medicine at Mount Sinai - Shreya Shah
Educator and Microbiologist, Cold Spring Harbor Laboratory
Finalists
Utilizing DNA Barcoding to Identify Macroinvertebrate Bioindicators in Queens Parks
Armani Khan and Jessica Kuppan, mentored by Karen Wong, High School For Construction Trades, Engineering, and Architecture
Proof of Panax: Discovering fraudulence in American ginseng products using DNA barcoding
Ella Epstein and Laura Glesby, mentored by Ileana Rios, Trinity School
The Effect of Human Impact On Invasive And Native Species Within A Marine Ecosystem
Daval Ram, Nick Holt, and Ron Bepat, mentored by Karen Wong, High School For Construction Trades, Engineering, and Architecture
Fraud, Mislabeling, and Misidentification of Fish Species Sold in New York City
Zachary Glass and Paul Kasiuaunis, mentored by Anat Firnberg, Tenafly High School
Bacteria in New York Water Sources
Sarafina Oh, Benjamn Shapiro, Caitlyn Tien, mentored by Matthew Wallenfang and Michelle Lee, Horace Mann School
Using DNA Barcode to Survey Seed Distribution Along the Bronx River
Josiah Estacio, Hayford Frempong, Laura Velazquez, and Lizbeth Ortigoza-Pacheco, mentored by Allison Granberry, Hostos-Lincoln Academy and Damon Little, New York Botanical Garden
Testing the Correlation Between the Bacterial Microbiomes Found on Mother’s and Their Children’s Smartphones
Azeez Adeyemi, Daniela Finkel, and Tyler Lederer-Plaskett, mentored by Howard Waldman and Adriana Andrade, Ethical Culture Fieldston School
2013-14 Symposium
Thanks to all of the teams who submitted proposals, performed experiments, and presented project results at the poster sessions held at the American Museum of Natural History on May 27, 2014. Six teams were selected to give oral presentations and compete for the grand prize at the UBP Symposium Award Ceremony on June 2, 2014 at AMNH.
At a glance:
- 38 participating teams
- 120 students from 26 high schools
- 22 mentors from 19 institutions
Taxonomic group studied:
- 47% plants
- 48% animals
- 5% fungi
Students submitted 500+ samples—an average of 13 samples/team —that generated over 800+ sequences.
Keynote & Finalist Presentations
- Welcome:
[00:00:00] Welcome
[00:03:00] Diversity of Ants in Park - Award Ceremony Remarks:
[00:07:05] Keynote – Mark Stoeckle, Ph.D. - Finalist Presentations:
[00:25:45] Hot Dogs: The Animal Inside
[00:37:30] Seahorses
[00:51:20] What's in Your Cup of Coffee
[01:06:20] Meat Industry Mislabeling
[01:17:05] Ethnic Market Sausage Mislabeling
[01:27:25] Halal Cart Meat Legitimacy<
Presentation of Awards
Judges:
- Mark Stoeckle, Ph.D.
Senior Research Associate, Program for the Human Environment, The Rockefeller University - Cristina Trowbridge
Senior Manager of Professional Development, American Museum of Natural History - David Kizirian, Ph.D.
Curatorial Associate, American Museum of Natural History
Finalists

Using DNA Barcoding to Compare the Trade of Traditional Chinese Medicine of Seahorses, and the Domestic Trade of Seahorses
Eric Bovee and Megan Peters, mentored by Marissa Bellino and J.T. Boehm, High School for Environmental Studies

Street Meat: Investigation of the Legitimacy of NYC Halal Carts Using DNA Barcoding
Maliha Rahman and Annie Yang, mentored by Ileana Rios, Trinity School
Nice to Meat You: Using DNA Barcoding to Detect Mislabeling in the Meat Industry
Sabrina Carrozzi, Grace McKenney, and Alexa Granser, mentored by Mary Musolino, Convent of the Sacred Heart
Using DNA barcodes to determine relationships between roasted and unroasted coffee beans cultivated in different parts of the world
Sabastian Rahman and Kailey Singh, mentored by Stacy Goldstein, Scholars Academy

An Investigation of Sausage Mislabeling in New York City Ethnic Markets
James Xu and Andrew Giordano, mentored by Ileana Rios, Trinity School
Hot Dogs, The Animal Inside: DNA Barcodes of Ten Different Hot Dog Brands
Sarafina Oh and Michael Wang, mentored by Matthew Wallenfang, Horace Mann School
2012-13 Symposium
Thanks to all of the teams who submitted proposals, performed experiments, and presented project results at the poster sessions held at the American Museum of Natural History on May 29, 2013. Six teams were selected to give oral presentations and compete for the grand prize at the UBP Symposium Award Ceremony on June 5, 2013 at AMNH. Funded by Alfred P. Sloan Foundation.
- 41 teams competing
- 113 students from 19 high schools
- 20 mentors from 14 institutions
- 54% animals
- 41% plants
- 5% fungi
More than 1,100 sequences, from approximately 600 samples. 15 samples per team on average
Keynote & Finalist Presentations
- Welcome:
[00:00:00] David Micklos - Award Ceremony Remarks:
[00:06:40] Keynote – George Amato, Ph.D. - Finalist Presentations:
[00:30:20] Birds Nest Soup
[00:42:50] Barcoding Snapping Turtles
[00:55:00] Allergy Causing Plants
[01:06:25] Pipefish in New York
[01:17:15] Ant Biodiversity
[01:28:50] Terrestrial Plant Bio-Indicators
Presentation of Awards
Judges:
- George Amato, Ph.D.
Director, Sackler Institute of Comparative Genomics, American Museum of Natural History - David Micklos
Executive Director, DNA Learning Center, Cold Spring Harbor Laboratory - Susan Perkins, Ph.D.
Curator, American Museum of Natural History
Finalists
Using DNA Barcodes to Identify Ant Biodiversity in St. Mary’s Park, Bronx, New York
Hillary Ramirez and Kavita Bhikhi, mentored by Allison Granberry, Hostos-Lincoln Academy

A Study of the Primary Ingredient in Birds Nest Soup, Aerodramus fuciphagus
Justin He and Eric Chen, mentored by Ileana Rios, Trinity School
Using Terrestrial Plant Bio-Indicators to Diagnose the Health of New York City
Victoria Majarali and Simranpreet Kanith, mentored by Kelly Lovelett, Brooklyn Technical High School
Pipefish in New York
Isaac Burg, Bard High School Early College Manhattan; Sophie Dornbaum, Brooklyn Technical High School; and Valerie Shoates, the Brearley School; mentored by Noah Burg, City University of New York, and Nuala Caomhánach, University College Dublin and AMNH
No More Itching and No More Ditching: Using DNA Barcoding to Identify Allergy Causing Plants
Faygie Feiner, Michal Leibowitz, Miriam Rosen, and Mindy Schwartz, mentored by Shulamith Biderman, Yeshiva University High School for Girls
DNA Barcoding Snapping Turtles (Chelydra serpentina) and Their Leech Parasites at Staten Island Parks
Kevin Lin and Akash Vaidya, mentored by Eugenia Naro-Maciel and Jenna Pantophlet, Staten Island Technical High School and the College of Staten Island, CUNY
2011-12 Symposium
Thanks to all of the teams who submitted proposals, performed experiments, and presented project results at the poster sessions held at the American Museum of Natural History on May 31 and June 1, 2012. Ten finalist teams presented projects at the Symposium on Wednesday, June 6, 2012. Funded by Alfred P. Sloan Foundation.
At a glance:
- 75 teams competing
- 218 students from 31 high schools, 23 public and 8 private
- 38 mentors from 29 institutions
Taxonomic group studied:
- 56% animals
- 36% plants
- 7% fungi
- 1% multiorganisms
More than 2,500 sequences, from approximately 1,000 samples. 15 samples per team on average
Award Ceremony Remarks
Rob DeSalle, Ph.D.
Curator, American Museum of Natural History
Rob DeSalle welcomes the student finalists and congratulates them on the importance of their projects to wildlife conservation.
Jesse H. Ausubel
Vice President, Alfred P. Sloan Foundation
Jesse H. Ausubel describes the history of DNA barcoding and congratulates the student teams on the diversity of their projects.
Bruce W. Stillman, Ph.D.
President, Cold Spring Harbor Laboratory
Bruce W. Stillman describes the history of the DNA Learning Center, and the unique experience provided by the Urban Barcode Project
Finalists
Traditional Chinese Medicine: Using Barcodes to Identify the Content of Ginkgo Products
Bobby Glover, Mary Acheampong, and Marisa VanBrakle, mentored by Allison Granberry Hostos-Lincoln Academy of Science in the Bronx
The Fungus amungus: DNA barcode-based survey of fungal biodiversity in Central Park
Tyler Bell, Elias Strizower, Sarah Sutto-Plunz, and Philip To, mentored by Marisa Wagner, The Bronx High School of Science
What species are sold as "catfish" in New York City?
Jalwa Afroz, Townsend Harris High School; with Wajiha Kazmi and Janki Tailor, Queens High School for the Sciences at York College; mentored by Elizabeth Alter, CUNY York College
Assessing Genetic Differences Among Atlantic Silversides (Menidia menidia) in New York
Caitlin Bauer, The Beacon School with Samantha Eng and Kadeem Walsh, Brooklyn Technical High School; mentored by Noah Burg, The American Museum of Natural History
Assessing the Identification, Distribution, and Diversity of Killifish in the Genus Fundulus in New York City Coastal Waters
Jeffrey Lin, Staten Island Technical High School, and Michelle Yu, The Bronx High School of Science, mentored by Noah Burg, The American Museum of Natural History
Barcoding Cimex lectularius
Era Lame and Nuttha Siriwatanakul mentored by Marissa Bellino, High School for Environmental Studies
Fruit Express: DNA Barcodes of Melons and Pears in Different Neighborhoods in NYC
Alissa Borshchenko, Jia Ling Chen, Lai Yee Kwan, and Whitney Lai, mentored by Kelly Lovelett, Brooklyn Technical High School
A Sea of Names: The Confusion Surrounding the Common Names of Fish
Ariela Farnham and August Zuzworsky, mentored by John Zuzworsky, Home-schooled students
South Bronx Ants: DNA Barcodes of Ants in St. Mary’s Park
Kavita Bhikhi, Lachoy Harris, Randol Mata, and Hillary Ramirez, mentored by Allison Granberry, Hostos-Lincoln Academy of Science
DNA Barcoding Exotic Agricultural Pests Seized by the U.S. Customs and Border Protection
Robyn Tse, Massapequa High School, mentored by Mark Stoeckle, Ph.D., The Rockefeller University